Task description¶
Task  ¶
¶
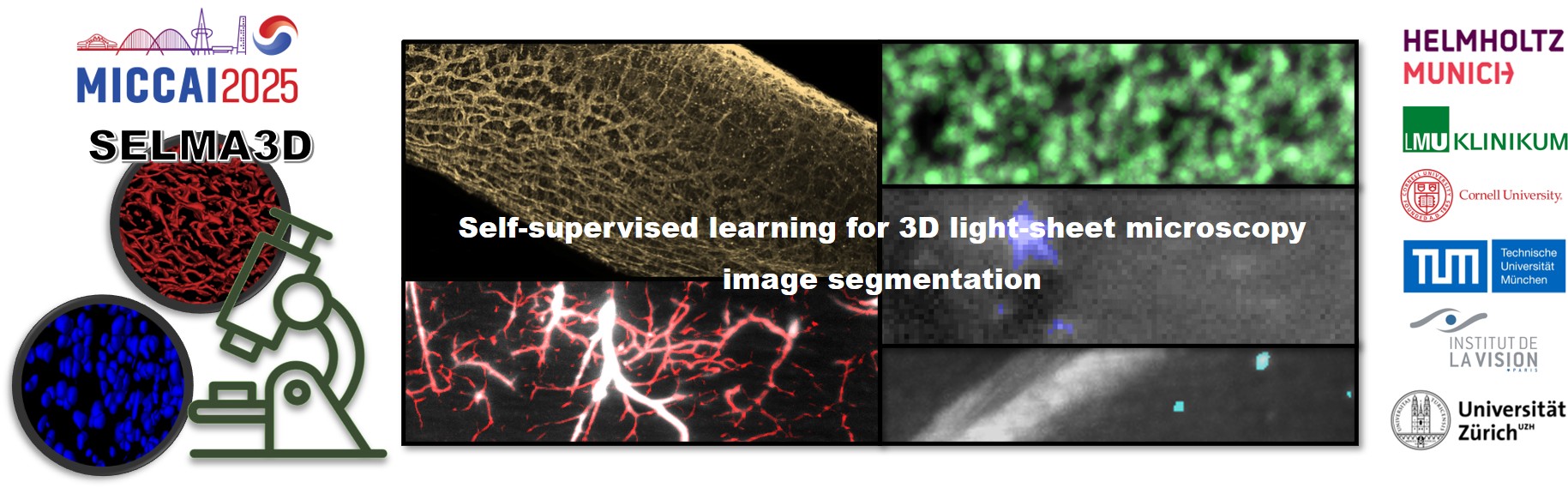
Task 1 is dedicated to developing SSL strategies for segmenting isolated structures in 3D light-sheet microscopy images. Isolated structures is one of the primary types of biological structures commonly studied in microscopy. These structures are characterized by spatially distinct components that lack physical continuity or direct connections. Examples include cell nuclei, specific cell types such as neurons or immune cells, and pathological formations like plaques (e.g., amyloid-beta plaques in Alzheimer’s disease).
The training dataset consists of two subsets:
- Unannotated subset: A large collection of 3D LSM images of isolated structures derived from both mouse and human samples. This subset includes 30 large high-resolution 3D images, each exceeding 2x10^10 voxels. These unannotated images are designed to support model pretraining using self-supervised learning techniques, enabling the model to learn generalizable representations of isolated structures.
- Annotated subset: A curated selection of 3D LSM image patches representing the same isolated structures in the unannotated subset, but with precise manual annotations. This subset enables participants to fine-tune their models for the segmentation of isolated structures.
Task  ¶
¶
Task 2 is dedicated to developing self-supervised learning strategies for segmenting contiguous structures in 3D light-sheet microscopy images. Contiguous structures represent another key type of biological structures commonly studied in microscopy. These structures are characterized by the physical continuity of their components, where different parts are connected without interruption, forming a continuous entity. Examples include blood vessels, lymphatic vessels, and nerves. In this task, participants will be provided with a training dataset consisting of two subsets:
- Unannotated subset: A large collection of 3D LSM images of contiguous structures derived from both mouse and human samples. This subset includes more than 25 high-resolution images, each exceeding 1x10^10 voxels. These unannotated images are designed to support model pretraining using self-supervised learning techniques, enabling the model to learn generalizable representations of contiguous structures.
- Annotated subset: A curated selection of 3D LSM image patches representing the same contiguous structures in the unannotated subset, but with precise manual annotations. This subset enables participants to fine-tune their models for the segmentation of contiguous structures.